Aragonites are Calcium carbonate crystals found in pearl.
In December 2009 issue of ProteinSpotlight, a very interesting protein is featured. This protein complex is none other than the one that gives luster to pearl. The complex is made out of three proteins, known as the Pif complex in which is found pearlin, Pif80 and Pif97. Pif80 and 97 are part of the same sequence which is subsequently cleaved into two.Pif80 – by way of its many asparagine residues – binds to calcium carbonate and not only elicits aragonite crystal formation but also has a role in the orientation of the aragonite crystals. Here is the Full link to the article..
Monday, December 21, 2009
Top five Biology papers for 2009
ScienceWatch tracks and analyzes trends in basic science research, compiles bimonthly lists of the 10 most cited papers. From that list The scientist pulled 5 papers published within the last two which were the most cited in 2009. The two topics that dominate the top five papers this year: genomics and stem cells.
Following is the list:
1. K. Takahashi, et al., "Induction of pluripotent stem cells from adult human fibroblasts by defined factors," Cell, 131: 861-72, 2007.
Citations this year: 520
Total citations to date: 886
Findings: This work from Shinya Yamanaka's lab in Japan was the first to demonstrate that induced pluripotent stem (iPS) cells can be generated from adult human dermal fibroblasts. Previous efforts by the team showed that iPS cells could be derived from mouse somatic cells. This paper was an easy top pick, receiving the most citations this year, according to ScienceWatch.
2. K.A. Frazer, et al., "A second generation human haplotype map of over 3.1 million SNPs," Nature, 449: 854-61, 2007.
Citations this year: 389
Total citations to date: 588
Findings: Since the sequencing of the human genome in 2003, the International HapMap Project has explored single nucleotide polymorphisms (SNPs) -- differences in a single letter of the DNA -- to study how these small variations affect the development of diseases and the body's response to pathogens and drugs. HapMap I, the original report, placed one SNP at roughly every 5,000 DNA letters. The newest map, featured in this paper, sequenced an additional 2 million SNPs, increasing the map's resolution to one SNP per kilobase. The additional detail allows scientists to more closely investigate patterns in SNP differences, especially in hotspot regions, or concentrated stretches of DNA.
3. A. Barski, et al., "High-resolution profiling of histone methylations in the human genome," Cell, 129: 823-37, 2007.
Citations this year: 299
Total citations to date:: 560
Findings: This study looked at how histone modifications influence gene expression in more detail than previous attempts. Using a powerful sequencing tool called Solexa 1G, the researchers mapped more than 20 million DNA sequences associated with specific forms of histones, finding there were differences in methylation patterns between stem cells and differentiated T cells.
4. E. Birney, et al., "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project, "Nature, 447: 799-816, 2007.
Citations this year: 267
Total citations to date: 618
Findings: The ENCODE project -- ENCODE stands for the ENCyclopedia Of DNA Elements -- set out to identify all functional elements in the human genome. After examining one percent of the genome, the paper revealed several new insights about how information encoded in the DNA comes to life in a cell.
5. A M. Wernig, et al., "In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state," Nature 448: 318-24, 2007.
Citations this year: 237
Total citations to date: 512
Findings: Scientists successfully performed somatic-cell nuclear transfer (SCNT), producing stem cell lines and cloned animals for the first time using fertilized mouse eggs. This paper consistently ranked in the top 10 most cited papers in 2009, according to ScienceWatch.
Following is the list:
1. K. Takahashi, et al., "Induction of pluripotent stem cells from adult human fibroblasts by defined factors," Cell, 131: 861-72, 2007.
Citations this year: 520
Total citations to date: 886
Findings: This work from Shinya Yamanaka's lab in Japan was the first to demonstrate that induced pluripotent stem (iPS) cells can be generated from adult human dermal fibroblasts. Previous efforts by the team showed that iPS cells could be derived from mouse somatic cells. This paper was an easy top pick, receiving the most citations this year, according to ScienceWatch.
2. K.A. Frazer, et al., "A second generation human haplotype map of over 3.1 million SNPs," Nature, 449: 854-61, 2007.
Citations this year: 389
Total citations to date: 588
Findings: Since the sequencing of the human genome in 2003, the International HapMap Project has explored single nucleotide polymorphisms (SNPs) -- differences in a single letter of the DNA -- to study how these small variations affect the development of diseases and the body's response to pathogens and drugs. HapMap I, the original report, placed one SNP at roughly every 5,000 DNA letters. The newest map, featured in this paper, sequenced an additional 2 million SNPs, increasing the map's resolution to one SNP per kilobase. The additional detail allows scientists to more closely investigate patterns in SNP differences, especially in hotspot regions, or concentrated stretches of DNA.
3. A. Barski, et al., "High-resolution profiling of histone methylations in the human genome," Cell, 129: 823-37, 2007.
Citations this year: 299
Total citations to date:: 560
Findings: This study looked at how histone modifications influence gene expression in more detail than previous attempts. Using a powerful sequencing tool called Solexa 1G, the researchers mapped more than 20 million DNA sequences associated with specific forms of histones, finding there were differences in methylation patterns between stem cells and differentiated T cells.
4. E. Birney, et al., "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project, "Nature, 447: 799-816, 2007.
Citations this year: 267
Total citations to date: 618
Findings: The ENCODE project -- ENCODE stands for the ENCyclopedia Of DNA Elements -- set out to identify all functional elements in the human genome. After examining one percent of the genome, the paper revealed several new insights about how information encoded in the DNA comes to life in a cell.
5. A M. Wernig, et al., "In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state," Nature 448: 318-24, 2007.
Citations this year: 237
Total citations to date: 512
Findings: Scientists successfully performed somatic-cell nuclear transfer (SCNT), producing stem cell lines and cloned animals for the first time using fertilized mouse eggs. This paper consistently ranked in the top 10 most cited papers in 2009, according to ScienceWatch.
Thursday, October 15, 2009
Death of End Note
I guess now End Note software will die a natural death!! I am not saying this just because I am not particularly fond of this software, but lately it has fierce competitions from various resources including my NCBI. Before My NCBI came into picture, I was very fond of connotea. Connotea is very easy to link to a browser, all that you will need is to drag and drop it to the menu bar. You can create your own login and password and update all the bibliography you ever wanted. Export the bibliography as you wish.
With My NCBI, it has struck the nail right on the head of End Note. Tutorials are readily available for My NCBI for easy use. Anytime, I would prefer a browser based application than a stand alone application. End note particularly needed endless filling up forms very tedious ways. I had bad experience of interference of end note with MS office 2007. Now I will breath easy writing a manuscript.
With My NCBI, it has struck the nail right on the head of End Note. Tutorials are readily available for My NCBI for easy use. Anytime, I would prefer a browser based application than a stand alone application. End note particularly needed endless filling up forms very tedious ways. I had bad experience of interference of end note with MS office 2007. Now I will breath easy writing a manuscript.
Monday, October 05, 2009
This years Nobel Prize in Medicine
This years Nobel prize in Medicine goes jointly to Elizabeth Blackburn, Carol Greider, and Jack Szostak for their work on telomeres. In the 1970s, Blackburn identified repeating segments at the ends of DNA in Tetrahymena while Szostak found that single-stranded DNA was rapidly degraded in yeast. Blackburn and Szostak then collaborated on a project, finding that the Tetrahymena DNA protected the single-stranded DNA from degradation in yeast. In 1984, Blackburn and Greider, her graduate student, discovered telomerase. The Nobel citation lauds these researchers for their contribution to the study of aging, cancer, and other diseases. "The discoveries by Blackburn, Greider and Szostak have added a new dimension to our understanding of the cell, shed light on disease mechanisms, and stimulated the development of potential new therapies," it says.
The mysterious telomere
The chromosomes contain our genome in their DNA molecules. As early as the 1930s, Hermann Muller (Nobel Prize 1946) and Barbara McClintock (Nobel Prize 1983) had observed that the structures at the ends of the chromosomes, the so-called telomeres, seemed to prevent the chromosomes from attaching to each other. They suspected that the telomeres could have a protective role, but how they operate remained an enigma.
When scientists began to understand how genes are copied, in the 1950s, another problem presented itself. When a cell is about to divide, the DNA molecules, which contain the four bases that form the genetic code, are copied, base by base, by DNA polymerase enzymes. However, for one of the two DNA strands, a problem exists in that the very end of the strand cannot be copied. Therefore, the chromosomes should be shortened every time a cell divides – but in fact that is not usually the case (Fig 1).
Both these problems were solved when this year's Nobel Laureates discovered how the telomere functions and found the enzyme that copies it.
Telomere DNA protects the chromosomes
In the early phase of her research career, Elizabeth Blackburn mapped DNA sequences. When studying the chromosomes of Tetrahymena, a unicellular ciliate organism, she identified a DNA sequence that was repeated several times at the ends of the chromosomes. The function of this sequence, CCCCAA, was unclear. At the same time, Jack Szostak had made the observation that a linear DNA molecule, a type of minichromosome, is rapidly degraded when introduced into yeast cells.
Blackburn presented her results at a conference in 1980. They caught Jack Szostak's interest and he and Blackburn decided to perform an experiment that would cross the boundaries between very distant species (Fig 2). From the DNA of Tetrahymena, Blackburn isolated the CCCCAA sequence. Szostak coupled it to the minichromosomes and put them back into yeast cells. The results, which were published in 1982, were striking – the telomere DNA sequence protected the minichromosomes from degradation. As telomere DNA from one organism, Tetrahymena, protected chromosomes in an entirely different one, yeast, this demonstrated the existence of a previously unrecognized fundamental mechanism. Later on, it became evident that telomere DNA with its characteristic sequence is present in most plants and animals, from amoeba to man.
An enzyme that builds telomeres
Carol Greider, then a graduate student, and her supervisor Blackburn started to investigate if the formation of telomere DNA could be due to an unknown enzyme. On Christmas Day, 1984, Greider discovered signs of enzymatic activity in a cell extract. Greider and Blackburn named the enzyme telomerase, purified it, and showed that it consists of RNA as well as protein (Fig 3). The RNA component turned out to contain the CCCCAA sequence. It serves as the template when the telomere is built, while the protein component is required for the construction work, i.e. the enzymatic activity. Telomerase extends telomere DNA, providing a platform that enables DNA polymerases to copy the entire length of the chromosome without missing the very end portion.
Blackburn was also the Daily Scan poll favorite. She led the pack with 43 percent of the vote. She was followed by her co-laureate Szostak who garnered 25 percent of the vote.
References:
Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid vectors. Cell 1982; 29:245-255.
Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985; 43:405-13.
Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989; 337:331-7.
The mysterious telomere
The chromosomes contain our genome in their DNA molecules. As early as the 1930s, Hermann Muller (Nobel Prize 1946) and Barbara McClintock (Nobel Prize 1983) had observed that the structures at the ends of the chromosomes, the so-called telomeres, seemed to prevent the chromosomes from attaching to each other. They suspected that the telomeres could have a protective role, but how they operate remained an enigma.
When scientists began to understand how genes are copied, in the 1950s, another problem presented itself. When a cell is about to divide, the DNA molecules, which contain the four bases that form the genetic code, are copied, base by base, by DNA polymerase enzymes. However, for one of the two DNA strands, a problem exists in that the very end of the strand cannot be copied. Therefore, the chromosomes should be shortened every time a cell divides – but in fact that is not usually the case (Fig 1).
Both these problems were solved when this year's Nobel Laureates discovered how the telomere functions and found the enzyme that copies it.
Telomere DNA protects the chromosomes
In the early phase of her research career, Elizabeth Blackburn mapped DNA sequences. When studying the chromosomes of Tetrahymena, a unicellular ciliate organism, she identified a DNA sequence that was repeated several times at the ends of the chromosomes. The function of this sequence, CCCCAA, was unclear. At the same time, Jack Szostak had made the observation that a linear DNA molecule, a type of minichromosome, is rapidly degraded when introduced into yeast cells.
Blackburn presented her results at a conference in 1980. They caught Jack Szostak's interest and he and Blackburn decided to perform an experiment that would cross the boundaries between very distant species (Fig 2). From the DNA of Tetrahymena, Blackburn isolated the CCCCAA sequence. Szostak coupled it to the minichromosomes and put them back into yeast cells. The results, which were published in 1982, were striking – the telomere DNA sequence protected the minichromosomes from degradation. As telomere DNA from one organism, Tetrahymena, protected chromosomes in an entirely different one, yeast, this demonstrated the existence of a previously unrecognized fundamental mechanism. Later on, it became evident that telomere DNA with its characteristic sequence is present in most plants and animals, from amoeba to man.
An enzyme that builds telomeres
Carol Greider, then a graduate student, and her supervisor Blackburn started to investigate if the formation of telomere DNA could be due to an unknown enzyme. On Christmas Day, 1984, Greider discovered signs of enzymatic activity in a cell extract. Greider and Blackburn named the enzyme telomerase, purified it, and showed that it consists of RNA as well as protein (Fig 3). The RNA component turned out to contain the CCCCAA sequence. It serves as the template when the telomere is built, while the protein component is required for the construction work, i.e. the enzymatic activity. Telomerase extends telomere DNA, providing a platform that enables DNA polymerases to copy the entire length of the chromosome without missing the very end portion.
Blackburn was also the Daily Scan poll favorite. She led the pack with 43 percent of the vote. She was followed by her co-laureate Szostak who garnered 25 percent of the vote.
References:
Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid vectors. Cell 1982; 29:245-255.
Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985; 43:405-13.
Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989; 337:331-7.
Friday, October 02, 2009
Oldest Human ancestor discovered
How old are humans? Until recently I thought Lucy to be our oldest ancestor, that is around 3.9 to 2.9 million years old. A recent finding by a group of scientists reveals our oldest known ancestor "Lucy" is now replaced by "Ardi"(Ardipithecus ramidus) which is older by 1.4 million years than Lucy.This individual, 'Ardi,' was a female who weighed about 50 kilograms and stood about 120 centimetres tall.
In its 2 October 2009 issue, Science presents 11 papers, authored by a diverse international team, describing an early hominid species, Ardipithecus ramidus, and its environment. These 4.4 million year old hominid fossils sit within a critical early part of human evolution, and cast new and sometimes surprising light on the evolution of human limbs and locomotion, the habitats occupied by early hominids, and the nature of our last common ancestor with chimps.
Science is making access to this extraordinary set of materials FREE (non-subscribers require a simple registration). The complete collection, and abridged versions, are available FREE as PDF downloads for AAAS members, or may be purchased as reprints.
The last common ancestor shared by humans and chimpanzees is thought to have lived six or more million years ago. Though Ardipithecus is not itself this last common ancestor, it likely shared many of this ancestor's characteristics. Ardi is closer to humans than chimps. Measuring in at 47 in. (120 cm) tall and 110 lb. (50 kg), Ardi likely walked with a strange gait, lurching side to side, due to lack of an arch in its feet, a feature of later hominids. It had somewhat monkey-like feet, with opposable toes, but its feet were not flexible enough to grab onto vines or tree trunks like many monkeys -- rather they were good enough to provide extra support during quick walks along tree branches -- called palm walking.
Another surprise comes in Ardi's environment. Ardi lived in a lush grassy African woodland, with creatures such as colobus monkeys, baboons, elephants, spiral-horned antelopes, hyenas, shrews, hares, porcupines, bats, peacocks, doves, lovebirds, swifts and owls. Fig trees grew around much of the area, and it is speculated that much of Ardi's diet consisted of these figs.
The surprise about the environment is that it lays to rest the theory that hominids developed upright walking when Africa's woodland-grassland mix changed to grassy savanna. Under this now theory, hominids began standing and walking upright as a way of seeing predators over the tall grasses. The discovery of Ardi -- an earlier upright walker that lived in woodland -- greatly weakens this theory.
Scientists have theorized that Ardi may have formed human-like relationships with pairing between single males and females. Evidence of this is found in the male's teeth, which lack the long canines that gorillas and other non-monogamous apes use to battle for females. Describes Professor Lovejoy, "The male canine tooth is no longer projecting or sharp. It's no longer weaponry."
In its 2 October 2009 issue, Science presents 11 papers, authored by a diverse international team, describing an early hominid species, Ardipithecus ramidus, and its environment. These 4.4 million year old hominid fossils sit within a critical early part of human evolution, and cast new and sometimes surprising light on the evolution of human limbs and locomotion, the habitats occupied by early hominids, and the nature of our last common ancestor with chimps.
Science is making access to this extraordinary set of materials FREE (non-subscribers require a simple registration). The complete collection, and abridged versions, are available FREE as PDF downloads for AAAS members, or may be purchased as reprints.
The last common ancestor shared by humans and chimpanzees is thought to have lived six or more million years ago. Though Ardipithecus is not itself this last common ancestor, it likely shared many of this ancestor's characteristics. Ardi is closer to humans than chimps. Measuring in at 47 in. (120 cm) tall and 110 lb. (50 kg), Ardi likely walked with a strange gait, lurching side to side, due to lack of an arch in its feet, a feature of later hominids. It had somewhat monkey-like feet, with opposable toes, but its feet were not flexible enough to grab onto vines or tree trunks like many monkeys -- rather they were good enough to provide extra support during quick walks along tree branches -- called palm walking.
Another surprise comes in Ardi's environment. Ardi lived in a lush grassy African woodland, with creatures such as colobus monkeys, baboons, elephants, spiral-horned antelopes, hyenas, shrews, hares, porcupines, bats, peacocks, doves, lovebirds, swifts and owls. Fig trees grew around much of the area, and it is speculated that much of Ardi's diet consisted of these figs.
The surprise about the environment is that it lays to rest the theory that hominids developed upright walking when Africa's woodland-grassland mix changed to grassy savanna. Under this now theory, hominids began standing and walking upright as a way of seeing predators over the tall grasses. The discovery of Ardi -- an earlier upright walker that lived in woodland -- greatly weakens this theory.
Scientists have theorized that Ardi may have formed human-like relationships with pairing between single males and females. Evidence of this is found in the male's teeth, which lack the long canines that gorillas and other non-monogamous apes use to battle for females. Describes Professor Lovejoy, "The male canine tooth is no longer projecting or sharp. It's no longer weaponry."
Friday, September 04, 2009
Restoring your dell laptop to factory setup
I have a dell laptop that had become extremely slow. I was looking at the options of cleaning the resgistry, cleaning up temporary internet files and all other available suggestions on internet. However, this did not help much. My laptop became almost unusable!! My desperate searches in the internet led me to find something like "While booting your system hold your control and f10 key" or "While booting your system hold your control and f11 key" or "While booting your system hold your control and f12 key" to get to setup option. I tried with F10, but it did not work, so I left the rest of the options.
The best way to look for the right combination of keys is to get hold of your DELL manual that comes along with your installation. It is sometimes also called "Owners manual". Once you got hold of it look for the "solving problem" -> "Restoring your operating system" section. Inside this section, you will find specific instructions as to how to get back to your factory default mode. For example mine says the following:
To use PC Restore:
1 Turn on the computer.
During the boot process, a blue bar with www.dell.com appears at the top of the screen.
2 Immediately upon seeing the blue bar, press < Ctrl >< F11 >.
If you do not press < Ctrl >< F11 > in time, let the computer finish starting, and then restart
the computer again.
NOTICE: If you do not want to proceed with PC Restore, click Reboot in the following step.
3 On the next screen that appears, click Restore.
4 On the next screen, click Confirm.
The restore process takes approximately 6–10 minutes to complete.
5 When prompted, click Finish to reboot the computer.
NOTE: Do not manually shut down the computer. Click Finish and let the computer completely
reboot.
6 When prompted, click Yes.
The computer restarts. Because the computer is restored to its original operating state, the
screens that appear, such as the End User License Agreement, are the same ones that
appeared the first time the computer was turned on.
7 Click Next.
The System Restore screen appears and the computer restarts.
8 After the computer restarts, click OK.
And needless to mention this worked like charm!! I got the desirable result. My computer works like new!! Only trouble is, you would have lost all your data and any new installations. But I don't think this is a big price to pay for this great change in performance. Installing softwares as you need and backing up data is something does not take a lot of effort.. So happy restoring your system.
The best way to look for the right combination of keys is to get hold of your DELL manual that comes along with your installation. It is sometimes also called "Owners manual". Once you got hold of it look for the "solving problem" -> "Restoring your operating system" section. Inside this section, you will find specific instructions as to how to get back to your factory default mode. For example mine says the following:
To use PC Restore:
1 Turn on the computer.
During the boot process, a blue bar with www.dell.com appears at the top of the screen.
2 Immediately upon seeing the blue bar, press < Ctrl >< F11 >.
If you do not press < Ctrl >< F11 > in time, let the computer finish starting, and then restart
the computer again.
NOTICE: If you do not want to proceed with PC Restore, click Reboot in the following step.
3 On the next screen that appears, click Restore.
4 On the next screen, click Confirm.
The restore process takes approximately 6–10 minutes to complete.
5 When prompted, click Finish to reboot the computer.
NOTE: Do not manually shut down the computer. Click Finish and let the computer completely
reboot.
6 When prompted, click Yes.
The computer restarts. Because the computer is restored to its original operating state, the
screens that appear, such as the End User License Agreement, are the same ones that
appeared the first time the computer was turned on.
7 Click Next.
The System Restore screen appears and the computer restarts.
8 After the computer restarts, click OK.
And needless to mention this worked like charm!! I got the desirable result. My computer works like new!! Only trouble is, you would have lost all your data and any new installations. But I don't think this is a big price to pay for this great change in performance. Installing softwares as you need and backing up data is something does not take a lot of effort.. So happy restoring your system.
Tuesday, July 21, 2009
ncftp and wget
As a bioinformatics researcher, it often becomes imperative to download a large amount of data sets from various servers. The most frequent data download site is NCBI and/or EBI. While most of the raw data can be found at NCBI, EBI hosts something much more curated. One nightmare I often face is, updating interproscan database. The data files are something like 9GB and take a lot of time to download, often exceeding the data limit for our server that downloads it. The most irritating of all is when it times out or says "the message list is too long". The best way to handle this trouble could be by using "wget" or "ncftp".
It is very simple to use and is very user friendly. Wget is very reliable and robust. It is specially designed for the home network if you are working from home using an unreliable network. If a download does not complete due to a network problem, Wget will automatically try to continue the download from where it left off, and repeat this until the whole file has been retrieved. It was one of the first clients to make use of the then-new Range HTTP header to support this feature.
If you are using the http protocol with wget then the format will be something like this:
wget --no-check-certificate https://login:passwd@site-address//path
or
wget ftp://ftp.gnu.org/pub/gnu/wget/wget-latest.tar.gz
Or more command info can be found by doing "man wget".
While wget is really cool, ncftp is another ftp protocol, that is sometimes much better than other existing methods, if you must use a ftp protocol for data download. A typical ncftp command could be:
ncftp -u login -p pass ftp://ftp.hostname.edu
Then use get command to get the files of interest.
It is very simple to use and is very user friendly. Wget is very reliable and robust. It is specially designed for the home network if you are working from home using an unreliable network. If a download does not complete due to a network problem, Wget will automatically try to continue the download from where it left off, and repeat this until the whole file has been retrieved. It was one of the first clients to make use of the then-new Range HTTP header to support this feature.
If you are using the http protocol with wget then the format will be something like this:
wget --no-check-certificate https://login:passwd@site-address//path
or
wget ftp://ftp.gnu.org/pub/gnu/wget/wget-latest.tar.gz
Or more command info can be found by doing "man wget".
While wget is really cool, ncftp is another ftp protocol, that is sometimes much better than other existing methods, if you must use a ftp protocol for data download. A typical ncftp command could be:
ncftp -u login -p pass ftp://ftp.hostname.edu
Then use get command to get the files of interest.
Thursday, July 16, 2009
Finding difference between 2 files
In bioinformatics work, one often encounters something like finding difference between 2 sequence files or finding Union and intersection between 2 datasets. This type of operation often needs comparison of one file with the other.
The most common way to do this job is to loop over first file and then loop over second file finding the match, if found terminate the internal loop reset the file pointer and again move on to the second element in the outer loop. This operation need O(M * N) time space. If size of M and N are huge than the amount of time space requirement is humongous. Using a binary search may reduce the time by O(log2 M * N), but the price will be paid again in sorting the files. If the files are non numeric, then the additional burden of converting strings to numbers will add to the price tag. So, recently, I have found using hash tables for finding an element, instead of looping through a second time is a great substitute for binary search. The files I was trying to search against were each having 2 million records. So, in a serial search, the file would have iterated 2 * 2 miilion times, comparing words 4 million times and resetting file 2 million times. While I was running this using the serial version the program ran for 2 days and was still running. The second approach just few minutes to run. Check this out..
Code for serial search:
#!/usr/bin/perl -w
# This script will find the difference between two list files
# Here find the difference between FH1 - FH2
open FH1, $ARGV[0] or die "Can't open file for reading $!\n";
open FH2, $ARGV[1] or die "Can't open file for reading $!\n";
my $flag = 1;
while(){
$flag = 1;
my $toBeSubtracted = $_;
while(){
if($_ eq $toBeSubtracted){
$flag =0;
last;
}
}
if($flag == 1){
print $toBeSubtracted;
}
seek(FH2,0,0);
}
Code using hash table:
#!/usr/bin/perl -w
# Subtract FH - FH1
open FH, $ARGV[0] or die "Can't open the file to be subtracted$!\n";
open FH1, $ARGV[1] or die "Can't open the file to be subtracted$!\n";
my %hash1;
my %hash2;
while(){
my $tmp = $_;
chomp($tmp);
$hash1{$tmp} = 1;
}
close(FH);
while(){
my $tmp = $_;
chomp($tmp);
$tmp =~ s/\t.*//g;
$hash2{$tmp} = 1;
}
close(FH1);
foreach my $key(keys %hash1){
if(exists($hash2{$key})){
#do nothing
}
else {
print "$key\n";
}
}
The most common way to do this job is to loop over first file and then loop over second file finding the match, if found terminate the internal loop reset the file pointer and again move on to the second element in the outer loop. This operation need O(M * N) time space. If size of M and N are huge than the amount of time space requirement is humongous. Using a binary search may reduce the time by O(log2 M * N), but the price will be paid again in sorting the files. If the files are non numeric, then the additional burden of converting strings to numbers will add to the price tag. So, recently, I have found using hash tables for finding an element, instead of looping through a second time is a great substitute for binary search. The files I was trying to search against were each having 2 million records. So, in a serial search, the file would have iterated 2 * 2 miilion times, comparing words 4 million times and resetting file 2 million times. While I was running this using the serial version the program ran for 2 days and was still running. The second approach just few minutes to run. Check this out..
Code for serial search:
#!/usr/bin/perl -w
# This script will find the difference between two list files
# Here find the difference between FH1 - FH2
open FH1, $ARGV[0] or die "Can't open file for reading $!\n";
open FH2, $ARGV[1] or die "Can't open file for reading $!\n";
my $flag = 1;
while(
$flag = 1;
my $toBeSubtracted = $_;
while(
if($_ eq $toBeSubtracted){
$flag =0;
last;
}
}
if($flag == 1){
print $toBeSubtracted;
}
seek(FH2,0,0);
}
Code using hash table:
#!/usr/bin/perl -w
# Subtract FH - FH1
open FH, $ARGV[0] or die "Can't open the file to be subtracted$!\n";
open FH1, $ARGV[1] or die "Can't open the file to be subtracted$!\n";
my %hash1;
my %hash2;
while(
my $tmp = $_;
chomp($tmp);
$hash1{$tmp} = 1;
}
close(FH);
while(
my $tmp = $_;
chomp($tmp);
$tmp =~ s/\t.*//g;
$hash2{$tmp} = 1;
}
close(FH1);
foreach my $key(keys %hash1){
if(exists($hash2{$key})){
#do nothing
}
else {
print "$key\n";
}
}
Cool unix tips
I work with large scale genome sequences, so I often have to handle huge files. The files many times have some unwanted characters and formats that I try to correct using vim substitute command. Although very powerful, substitution operation on huge files using vim is very time consuming!!
A very powerful option to vim substitution is sed(stream editor) utility. This utility is often under utilized because of lack of proper documentation. Some of very powerful file editing can be done using sed.
Simple Substitution:
sed 's/\/usr\/local\/bin/\/common\/bin/'new # will replace /usr/local/bin by /common/bin
Gulp. Some call this a 'Picket Fence' and it's ugly. It is easier to read if you use an underline instead of a slash as a delimiter:
sed 's_/usr/local/bin_/common/bin_'new
Some people use colons:
sed 's:/usr/local/bin:/common/bin:'new
Others use the "|" character.
sed 's|/usr/local/bin|/common/bin|'new
Replacing a pattern with something else:
In a file if the content is something like: '123 abc' and you want to make it 123 123 abc, then do the following:
sed 's/[0-9]*/& &/' < old > new # Here '&' serves as the pattern
Meaning of \1 and \2 till \9
These simply mean the pattern number. Altogether sed can remember upto 9 patterns.
Suppose say in a file you would like to replace "1234 abcd" by "abcd 1234" then do the following;
sed 's/\([0-9]*\) \([a-z]*\)/\2 \1/'. Here notice the space between the two patterns.
Substituting only in some lime lines then use the following:
sed '101,532 s/A/a/' or sed '101,$ s/A/a/'
Deleting lines beginning with # is easy:
sed '/^#/ d'
This one will remove comments and blank lines:
sed -e 's/#.*//' -e '/^$/ d'
This one will remove all tabs and blanks before end of the line:
sed -e 's/#.*//' -e 's/[ ^I]*$//' -e '/^$/ d'
Before I sign off how to tell if your machine is 64 bit linux or 32 bit?
uname -m
i386 or i686 then it is 32 bit
x86_64 is 64bit
Getting cpu info:
more /proc/cpuinfo
Some more Generic unix shell scripts:
Total number of files in your working area: ls / -R | wc -l &
To see a process you are really interested in seeing: ps ax | grep httpd
To display a tree of processes: pstree
calculator program in unix: bc ( It will wait for your inputs)
Cool Small Applications:
Chessboard:
for (( i = 1; i <= 9; i++ )) ### Outer for loop ### do for (( j = 1 ; j <= 9; j++ )) ### Inner for loop ### do tot=`expr $i + $j` tmp=`expr $tot % 2` if [ $tmp -eq 0 ]; then echo -e -n "\033[47m " else echo -e -n "\033[40m " fi done echo -e -n "\033[40m" #### set back background colour to black echo "" #### print the new line ### done Use Cut and paste command
I find these 2 commands kind of blessing. cut will cut the columns of a flat file for you and paste will merge them together. There is one more command join that joins 2 files if they have a common field...
These utilities are extremely powerful if you are using them in merging 2 or more data files having background corrected/normalized data or something like that. Perl scripts are expansive in terms of file handling.
Using awk:
Did we know that the present day text editor perl burrows most of its syntax from unix shell scripting, particularly from awk.
awk is a powerful pattern matcher having most of its syntax similar to perl or should I say perl has similar syntax as awk.
awk '
BEGIN { actions }
/pattern/ { actions }
/pattern/ { actions }
END { actions }
' files
in the quoted area you can put anything like a condition such as
awk ‘{ if($5 !~ /[0-9]/ || NF != 7) {print “Line Number ” NR “ has error: ” $0}}’ Input file
This command will check if the input file has column 5 non-numeric value and if the input file has 7 fields or not. In case it is not, this is going to print the file having error. In case of no error it just prints nothing. NR is a special character meaning line number. NF stands for number of fields. By default awk takes [tab] as field separator. But in case you have something else as a separator you can begin awk with:
awk '
BEGIN { FS =":"; }
/pattern/ { actions }
/pattern/ { actions }
END { actions }
' files
For example if you are scanning a file for a particular pattern and you want to print that line having the pattern
awk ' BEGIN{FS = ":"; }
/pattern/ {i++; print "Line Number: " NR ":" $0}
END {printf("Number of lines having pattern %d\n",i)}
'
This command will print
Line Number : 1 : suydusydu 88 sllsilsdi
Line Number :5 : suyfdusydu 88 sllsilsdi
Line Number :6 : suycdusydu 88 sllsilsdi
Line Number :7 : suyvdusydu 88 sllsilsdi
Line Number :8 : suygdusydu 88 sllsilsdi
Line Number :9 : suydugsydu 88 sllsilsdi
Line Number :10 : suydusggydu 88 sllsilsdi
Line Number :11 : suydusygdu 88 sllsilsdi
Line Number :12 : suydusgydu 88 sllsilsdi
Line Number :13 : suydusffydu uiuiu sllsilsdi
Line Number :14 : suydusssydu 88 sllsilsdi
Line Number :15 : suydussydu 88 sllsilsdi
Line Number :16 : suydussssydu 88 sllsilsdi
Line Number :17 : suydusssssydu 88 sllsilsdi
Line Number :18 : suydusyssadu 88 sllsilsdi
Line Number :19 : suydusyqqdu 88 sllsilsdi
Line Number :20 : suydusydffu 88 sllsilsdi
Number of lines having pattern 17
A very powerful option to vim substitution is sed(stream editor) utility. This utility is often under utilized because of lack of proper documentation. Some of very powerful file editing can be done using sed.
Simple Substitution:
sed 's/\/usr\/local\/bin/\/common\/bin/'
Gulp. Some call this a 'Picket Fence' and it's ugly. It is easier to read if you use an underline instead of a slash as a delimiter:
sed 's_/usr/local/bin_/common/bin_'
Some people use colons:
sed 's:/usr/local/bin:/common/bin:'
Others use the "|" character.
sed 's|/usr/local/bin|/common/bin|'
Replacing a pattern with something else:
In a file if the content is something like: '123 abc' and you want to make it 123 123 abc, then do the following:
sed 's/[0-9]*/& &/' < old > new # Here '&' serves as the pattern
Meaning of \1 and \2 till \9
These simply mean the pattern number. Altogether sed can remember upto 9 patterns.
Suppose say in a file you would like to replace "1234 abcd" by "abcd 1234" then do the following;
sed 's/\([0-9]*\) \([a-z]*\)/\2 \1/'. Here notice the space between the two patterns.
Substituting only in some lime lines then use the following:
sed '101,532 s/A/a/' or sed '101,$ s/A/a/'
Deleting lines beginning with # is easy:
sed '/^#/ d'
This one will remove comments and blank lines:
sed -e 's/#.*//' -e '/^$/ d'
This one will remove all tabs and blanks before end of the line:
sed -e 's/#.*//' -e 's/[ ^I]*$//' -e '/^$/ d'
Before I sign off how to tell if your machine is 64 bit linux or 32 bit?
uname -m
i386 or i686 then it is 32 bit
x86_64 is 64bit
Getting cpu info:
more /proc/cpuinfo
Some more Generic unix shell scripts:
Total number of files in your working area: ls / -R | wc -l &
To see a process you are really interested in seeing: ps ax | grep httpd
To display a tree of processes: pstree
calculator program in unix: bc ( It will wait for your inputs)
Cool Small Applications:
Chessboard:
for (( i = 1; i <= 9; i++ )) ### Outer for loop ### do for (( j = 1 ; j <= 9; j++ )) ### Inner for loop ### do tot=`expr $i + $j` tmp=`expr $tot % 2` if [ $tmp -eq 0 ]; then echo -e -n "\033[47m " else echo -e -n "\033[40m " fi done echo -e -n "\033[40m" #### set back background colour to black echo "" #### print the new line ### done Use Cut and paste command
I find these 2 commands kind of blessing. cut will cut the columns of a flat file for you and paste will merge them together. There is one more command join that joins 2 files if they have a common field...
These utilities are extremely powerful if you are using them in merging 2 or more data files having background corrected/normalized data or something like that. Perl scripts are expansive in terms of file handling.
Using awk:
Did we know that the present day text editor perl burrows most of its syntax from unix shell scripting, particularly from awk.
awk is a powerful pattern matcher having most of its syntax similar to perl or should I say perl has similar syntax as awk.
awk '
BEGIN { actions }
/pattern/ { actions }
/pattern/ { actions }
END { actions }
' files
in the quoted area you can put anything like a condition such as
awk ‘{ if($5 !~ /[0-9]/ || NF != 7) {print “Line Number ” NR “ has error: ” $0}}’ Input file
This command will check if the input file has column 5 non-numeric value and if the input file has 7 fields or not. In case it is not, this is going to print the file having error. In case of no error it just prints nothing. NR is a special character meaning line number. NF stands for number of fields. By default awk takes [tab] as field separator. But in case you have something else as a separator you can begin awk with:
awk '
BEGIN { FS =":"; }
/pattern/ { actions }
/pattern/ { actions }
END { actions }
' files
For example if you are scanning a file for a particular pattern and you want to print that line having the pattern
awk ' BEGIN{FS = ":"; }
/pattern/ {i++; print "Line Number: " NR ":" $0}
END {printf("Number of lines having pattern %d\n",i)}
'
This command will print
Line Number : 1 : suydusydu 88 sllsilsdi
Line Number :5 : suyfdusydu 88 sllsilsdi
Line Number :6 : suycdusydu 88 sllsilsdi
Line Number :7 : suyvdusydu 88 sllsilsdi
Line Number :8 : suygdusydu 88 sllsilsdi
Line Number :9 : suydugsydu 88 sllsilsdi
Line Number :10 : suydusggydu 88 sllsilsdi
Line Number :11 : suydusygdu 88 sllsilsdi
Line Number :12 : suydusgydu 88 sllsilsdi
Line Number :13 : suydusffydu uiuiu sllsilsdi
Line Number :14 : suydusssydu 88 sllsilsdi
Line Number :15 : suydussydu 88 sllsilsdi
Line Number :16 : suydussssydu 88 sllsilsdi
Line Number :17 : suydusssssydu 88 sllsilsdi
Line Number :18 : suydusyssadu 88 sllsilsdi
Line Number :19 : suydusyqqdu 88 sllsilsdi
Line Number :20 : suydusydffu 88 sllsilsdi
Number of lines having pattern 17
Wednesday, July 01, 2009
DNA Methylation AIDS HIV Latency
Current drug therapies inhibit replication of the human immunodeficiency virus (HIV). In patients undergoing these therapies, the amount of HIV is reduced to an undetectable level and HIV-related disease subsides. However, stopping antiviral drug therapy results in the quick return of HIV and of disease. One reason for this is latently infected cells, in which virus replication is temporarily halted. When drug therapy is stopped, virus from these latently infected cells can resume infection and spread to other cells in the patient, resulting in the return of disease
One of the world's most elusive viruses is an expert at maintaining a low profile, laying dormant in CD4+ cells even during highly active anti-retroviral treatment (HAART). A team of American and Swedish researchers found that the virus might be using DNA methylation as a cloak.
Hypermethylated CpG islands flanking the HIV transcription site attract methyl-CpG binding domain protein 2 (MBD2) -- an endogenous host protein – which in turn recruits histone deacetylaces and other enzymes to shut down transcription.
Using 5-aza-deoxycytidine (Aza-CdR) to strip the DNA methylation at these island allowed researchers to reverse the transcriptional block and reactivate HIV right out of hiding, indicating that Aza-CdR might be a great complement to other antiviral therapies. So there’s hope for flushing out the reservoir, clearing patients of HIV-1, and letting them live a drug free life. Wouldn’t Nancy Reagan be proud?
See all the HAARTening details at PloS Pathogens June 2009.
One of the world's most elusive viruses is an expert at maintaining a low profile, laying dormant in CD4+ cells even during highly active anti-retroviral treatment (HAART). A team of American and Swedish researchers found that the virus might be using DNA methylation as a cloak.
Hypermethylated CpG islands flanking the HIV transcription site attract methyl-CpG binding domain protein 2 (MBD2) -- an endogenous host protein – which in turn recruits histone deacetylaces and other enzymes to shut down transcription.
Using 5-aza-deoxycytidine (Aza-CdR) to strip the DNA methylation at these island allowed researchers to reverse the transcriptional block and reactivate HIV right out of hiding, indicating that Aza-CdR might be a great complement to other antiviral therapies. So there’s hope for flushing out the reservoir, clearing patients of HIV-1, and letting them live a drug free life. Wouldn’t Nancy Reagan be proud?
See all the HAARTening details at PloS Pathogens June 2009.
Friday, April 03, 2009
This is a great article on miRNA
I read this article liked it a lot. So, copy pasting it for future reference:
http://www.nature.com/scitable/topicpage/Small-Non-coding-RNA-and-Gene-Expression-1078
http://www.nature.com/scitable/topicpage/Small-Non-coding-RNA-and-Gene-Expression-1078
Monday, March 30, 2009
Fungus farmers show way to new drugs
[I came across this interesting story in nature news, thought I will put it in my blog:]
Ant colonies could be key to advances in biofuels and antibiotics.
-Erika Check Hayden
Studies of bacteria on leaf-cutting ants could yield new antibiotics.Studies of bacteria on leaf-cutting ants could yield new antibiotics.M. MOFFETT/FLPA
In a mutually beneficial symbiosis, leaf-cutting ants cultivate fungus gardens, providing both a safe home for the fungi and a food source for the ants. But this 50-million-year-old relationship also includes microbes that new research shows could help speed the quest to develop better antibiotics and biofuels.
Ten years ago, Cameron Currie, a microbial ecologist then at the University of Toronto in Ontario, Canada, discovered that leaf-cutting ants carry colonies of actinomycete bacteria on their bodies (C. R. Currie et al. Nature 398, 701–704; 1999). The bacteria churn out an antibiotic that protects the ants' fungal crops from associated parasitic fungi (such as Escovopsis). On 29 March, Currie, Jon Clardy at the Harvard Medical School in Boston and their colleagues reported that they had isolated and purified one of these antifungals, which they named dentigerumycin, and that it is a chemical that has never been previously reported (D.-C. Oh et al. Nature Chem. Bio. doi: 10.1038/nchembio.159; 2009). The antifungal slowed the growth of a drug-resistant strain of the fungus Candida albicans, which causes yeast infections in people.
“These ants are walking pharmaceutical factories.”
Because distinct ant species cultivate different fungal crops, which in turn fall prey to specialized parasites, researchers hope that they will learn how to make better antibiotics by studying how the bacteria have adapted to fight the parasite in an ancient evolutionary arms race. "These ants are walking pharmaceutical factories," says Currie, now at the University of Wisconsin, Madison.
That's not the end to the possible applications. The ant colonies are also miniature biofuel reactors, Currie reported on 25 March at the Genomics of Energy & Environment meeting at the Joint Genome Institute in Walnut Creek, California. Each year, ants from a single colony harvest up to 400 kilograms of leaves to feed their fungal partners. But no one has worked out how the fungi digest the leaves, because samples of fungus grown in petri dishes can't break down cellulose, a tough molecule found in plant cells. Researchers are keenly interested in better ways to break down cellulose, because it might allow them to make more efficient biofuels than those made from sugary foods, such as maize (corn).
So Currie and his colleagues sequenced small segments of DNA from bacteria and other organisms living in fungus gardens in three Panamanian leaf-cutting ant colonies. They then compared the DNA against databases to identify what species were living in the fungus gardens, and what genes they contained.
This 'metagenomics' approach found that there are many species of bacteria in the fungus gardens that are capable of breaking down cellulose. The team also detected the genetic signatures of fungal enzymes that can break down cellulose, which raises the question of why the fungi can't break down cellulose in the laboratory.
Currie suggests that the newfound bacterial and fungal enzymes might be efficient at digesting cellulose because they have evolved for centuries along with the ant-fungal symbiosis. This could mean that the fungus can only break down cellulose in its natural context, or that the enzymes Currie detected are brought into the colony from outside. "The idea is that the ants' long evolutionary history may help us in our own attempts to break down plant biomass," he says.
Other researchers call Currie's findings interesting, but say they wanted to see a more thorough analysis of the data. "It's interesting that he found these fungal enzymes in the gardens that he didn't expect [based on] what the fungus was capable of doing by itself," says John Taylor, a mycologist at the University of California, Berkeley.
Taylor says that Currie's continued scrutiny of the lives of ants provides insights into the web of interactions necessary for the survival of any single species. "I think the coolest thing about this is that you start with one organism, and then you find more and more organisms involved in the relationship," he says. It may take a village to raise a child; it seems it also takes a village to break down cellulose.
Ant colonies could be key to advances in biofuels and antibiotics.
-Erika Check Hayden
Studies of bacteria on leaf-cutting ants could yield new antibiotics.Studies of bacteria on leaf-cutting ants could yield new antibiotics.M. MOFFETT/FLPA
In a mutually beneficial symbiosis, leaf-cutting ants cultivate fungus gardens, providing both a safe home for the fungi and a food source for the ants. But this 50-million-year-old relationship also includes microbes that new research shows could help speed the quest to develop better antibiotics and biofuels.
Ten years ago, Cameron Currie, a microbial ecologist then at the University of Toronto in Ontario, Canada, discovered that leaf-cutting ants carry colonies of actinomycete bacteria on their bodies (C. R. Currie et al. Nature 398, 701–704; 1999). The bacteria churn out an antibiotic that protects the ants' fungal crops from associated parasitic fungi (such as Escovopsis). On 29 March, Currie, Jon Clardy at the Harvard Medical School in Boston and their colleagues reported that they had isolated and purified one of these antifungals, which they named dentigerumycin, and that it is a chemical that has never been previously reported (D.-C. Oh et al. Nature Chem. Bio. doi: 10.1038/nchembio.159; 2009). The antifungal slowed the growth of a drug-resistant strain of the fungus Candida albicans, which causes yeast infections in people.
“These ants are walking pharmaceutical factories.”
Because distinct ant species cultivate different fungal crops, which in turn fall prey to specialized parasites, researchers hope that they will learn how to make better antibiotics by studying how the bacteria have adapted to fight the parasite in an ancient evolutionary arms race. "These ants are walking pharmaceutical factories," says Currie, now at the University of Wisconsin, Madison.
That's not the end to the possible applications. The ant colonies are also miniature biofuel reactors, Currie reported on 25 March at the Genomics of Energy & Environment meeting at the Joint Genome Institute in Walnut Creek, California. Each year, ants from a single colony harvest up to 400 kilograms of leaves to feed their fungal partners. But no one has worked out how the fungi digest the leaves, because samples of fungus grown in petri dishes can't break down cellulose, a tough molecule found in plant cells. Researchers are keenly interested in better ways to break down cellulose, because it might allow them to make more efficient biofuels than those made from sugary foods, such as maize (corn).
So Currie and his colleagues sequenced small segments of DNA from bacteria and other organisms living in fungus gardens in three Panamanian leaf-cutting ant colonies. They then compared the DNA against databases to identify what species were living in the fungus gardens, and what genes they contained.
This 'metagenomics' approach found that there are many species of bacteria in the fungus gardens that are capable of breaking down cellulose. The team also detected the genetic signatures of fungal enzymes that can break down cellulose, which raises the question of why the fungi can't break down cellulose in the laboratory.
Currie suggests that the newfound bacterial and fungal enzymes might be efficient at digesting cellulose because they have evolved for centuries along with the ant-fungal symbiosis. This could mean that the fungus can only break down cellulose in its natural context, or that the enzymes Currie detected are brought into the colony from outside. "The idea is that the ants' long evolutionary history may help us in our own attempts to break down plant biomass," he says.
Other researchers call Currie's findings interesting, but say they wanted to see a more thorough analysis of the data. "It's interesting that he found these fungal enzymes in the gardens that he didn't expect [based on] what the fungus was capable of doing by itself," says John Taylor, a mycologist at the University of California, Berkeley.
Taylor says that Currie's continued scrutiny of the lives of ants provides insights into the web of interactions necessary for the survival of any single species. "I think the coolest thing about this is that you start with one organism, and then you find more and more organisms involved in the relationship," he says. It may take a village to raise a child; it seems it also takes a village to break down cellulose.
Thursday, March 26, 2009
Using galaxy server
I recently came across galaxy server for genome sequence manipulation. While I do large scale computing, I myself have thought several times to create a web based tool for large scale sequence manipulation, that will be able to handle huge data files. My requirements would often be to subtract sequences from one file to another, fetch sequences from huge fasta file containing names from a list file, find duplicate triplicate sequences, find sequences of a certain length etc. I think the galaxy server provides many of that through a web server. The link is here
creating a wiki page for community portal
I have been toying with the idea of creating a wiki for our VMD web portal. The rationale is to create a user document in a wiki, which will also be used for bug tracking versioning etc. I am now looking at freely available tools/softwares for wiki creation( a compiled version is available here ).
For me documentation in wiki could be a great idea, since users can edit this page at ease and complement each others observation, instead of one person writing a whole document. Docuwiki seem to be the simplest one without the intervention of database. It just handles files and so I would assume is much safer option for our servers.
For me documentation in wiki could be a great idea, since users can edit this page at ease and complement each others observation, instead of one person writing a whole document. Docuwiki seem to be the simplest one without the intervention of database. It just handles files and so I would assume is much safer option for our servers.
Monday, March 09, 2009
Find pattern using grep (egrep)
One of the quickest and easiest ways to find patterns in a huge file without writing any scripts is through unix grep. I find it extremely useful to fetch strings having a certain pattern.
Looking for Tabs:
Many manuals don't say a lot about finding a tab character in a pattern. If you have a pattern "gene TAB exon", then the way to look for the file would be:
grep 'gene[[:blank:]]exon'
Looking for Tabs:
Many manuals don't say a lot about finding a tab character in a pattern. If you have a pattern "gene TAB exon", then the way to look for the file would be:
grep 'gene[[:blank:]]exon'
Monday, March 02, 2009
Implementing binary search in searching a list of sequences from a complex file
In sequence analysis work, it becomes imperative to search a list of sequences from a sequence file or a file having complex sets of data. One such complex dataset I can think of is an ACE file. An ACE file has multiple components:
Suppose you have a list of sequences in a file and you got to search which sequence belongs to which Contig from the ACE file. Then probably, you have to read the ACE file first as an hash of arrays, something like this:
After reading ACE into hash of arrays, need to iterate over the hash and get the sequence of arrays searched against each entry in the file. If the sequence file size is hugh, its good to implement a binary search(O(log2N)) instead of a serial search(O(N)). But knowing the fact that binary search can be implemented on a sorted array or list, it is more probable to sort the list file instead of sorting the array of hash. Because once sorted the list file stays sorted and does not need any sorting.
Implementation:
my @list = sort(@list); # sorting the sequence names in list file
AS contigs reads
CO contig_name bases reads segments compl (CAP3: segments=0)
sequence
BQ base_qualities
AF read1 compl padded_start_consensus (negatives meaning?)
AF read2 ..
BS segments
RD read1 bases info_items info_tags (latter two set to 0 by CAP3)
sequence
QA read1 qual_start qual_end align_start align_end
DS (phred header? left empty by CAP3)
RD read2 ...
Suppose you have a list of sequences in a file and you got to search which sequence belongs to which Contig from the ACE file. Then probably, you have to read the ACE file first as an hash of arrays, something like this:
while(){
if(/^CO Contig/){
$key = $_;
$flag = 1;
}
if($flag && /^[ATGC][ATGC][ATGC]/){
chomp;
$seq.=$_;
}
if(/^BQ/){
push(@arr,$seq);
$seq = '';
}
if(/^AF\s+(\S+)/){
push(@arr,$1);
}
if(/^BS/ && $flag){
$HOA{$key} = [ @arr ];
$flag = 0;
@arr = ();
}
}
After reading ACE into hash of arrays, need to iterate over the hash and get the sequence of arrays searched against each entry in the file. If the sequence file size is hugh, its good to implement a binary search(O(log2N)) instead of a serial search(O(N)). But knowing the fact that binary search can be implemented on a sorted array or list, it is more probable to sort the list file instead of sorting the array of hash. Because once sorted the list file stays sorted and does not need any sorting.
Implementation:
my @list = sort(@list); # sorting the sequence names in list file
# The following implementation is a linear search and takes
# much longer time
foreach my $key(keys %HOA){
my @arr = @{ $HOA{$key}} ;
my @found;
for(my $i=1;$i<=$#arr;$i++){
for(my $j=0;$j<=$#list;$j++){
chomp($list[$j]);
if($arr[$i] =~ $list[$j]){
push(@found, $arr[$i]);
splice(@list,$j,1); # removing the
# element from the list
last;
}
}
}
Binary search:
for(my $i=1;$i<=$#arr;$i++){
push(@found, bsearch($arr[$i],\@list);
}
sub bsearch {
my ($x, $a) = @_; # search for x in array a
my ($l, $u) = (0, @$a - 1); # lower, upper end of search interval
my $i; # index of probe
while ($l <= $u) {
$i = int(($l + $u)/2);
print($i, "\n");
if ($a->[$i] < $x) {
$l = $i+1;
}
elsif ($a->[$i] > $x) {
$u = $i-1;
}
else {
return $i; # found
}
}
return -1; # not found
}
Thursday, February 19, 2009
Cool perl tips
While dealing with very huge files, it is often necessary not to read the entire file into an array. This will be a huge RAM cost to the machine. Iterating through a file handle is often a easy solution. But often we need to go back and forth the file as we find a match. The easiest way to achieve this will be to use perl 'seek' function.
Similarly, Redo operator can also be explored to move back in loop.
Searching patterns in 2 data files:
In bioinformatics work, it is often necessary to search a list of names from a file from another file, that either has a list of sequences or blast output or anything else. So, the trick to reduce the search time will be keep the larger file as the outer loop(mostly as a while loop), the inside loop could be a while loop or a for loop, depending on the file size. If there is a for loop for the inner loop, then it is possible to get rid of the element that has just had a match to the outer element. This way with every iteration, the inner loop gets shorter and the time spent in searching gets shorter. In perl it is quite easy to remove an array element by simply using a delete function.
delete($array[index]) will do the job. But just delete removes the element while leaving $array[index] space empty. In order to remove the element along with index use splice function instead.
splice(@array,index,offset) --> will remove offset number of elements and indices starting from index. i.e; splice(@array,2,7) will remove 7 indices starting from 2.
The whole construct may look something like this:
If you have a subroutine and you need to pass more than one array, then don't pass by value. If passed by value then the list is flattened and the first and second element of the array is passed as 2 arrays. Example: @arr1 = [1,2,3];
@arr2 = [6,7,8];
func(@arr1,@arr2);
will print: Array1 is 1 and array2 is 2
sub func
{
my @a = shift;
my @b = shift;
print "Array1 is @a and array2 is @b\n";
}
In order to get the values intact pass by reference:
func(\@arr1,\@arr2);
in
sub func{
$a = shift;
$b = shift;
@arr1 = @$a; # de-referencing array
@arr2 = @$b;
Here is a time and space saver if you want to sort a hash by value and iterate over the sorted list:
# sorting tmp hash by value
foreach my $sorted(sort {$tmp{$b} <=> $tmp{$a}} keys %tmp){
my %geneHash;
for my $gene(keys %{$HOH1{$sorted}}){
my @coord = sort{$a <=> $b} @{ $HOH1{$sorted}{$gene}{'pos'}};
my $strand1 = $HOH1{$sorted}{$gene}{'strand'};
$geneHash{$gene} = $coord[0];
}
foreach my $sortedgene(sort {$geneHash{$a} <=> $geneHash{$b}} keys %geneHash ){
print FH "$sortedgene\t$serielNumber\n";
print FH1 ">
$serielNumber\n";
print FH1 "$transcript{$sortedgene}\n";
`sed 's/\t$sortedgene/$serielNumber/' output.gff`;
$serielNumber++;
}
}
# One variation if you want to sort the sequence file in descending order of their length. step -1:
Read fasta file into a hash called as %hash
# sort hash by the length of the value
foreach my $scaffold(sort {length($hash{$b}) <=> length($hash{$a}) } keys %hash) {
print ">scaffold_$num\n$hash{$scaffold}\n";
print FH "$scaffold\tscaffold_$num\t".length($hash{$scaffold})."\n"; $num++;
}
To make it to the ascending order simply change this to:
foreach my $scaffold(sort {length($hash{$a}) <=> length($hash{$b}) } keys %hash)
while(){ some pattern.. seek(FH, - length($_), 1); # will point the File handle # one line back }
NOTE: Just reading one line of a file at a time can be done by simply;
$_= ; # where FH stands for file handle.
Next call to $_= ; will read the next line. This is a great way to read
2 files together especially if they both are representing sequence and qual files
and the number of lines and values are in same order in both the files. Simply iterate
one file with a while loop and call $_= inside the while loop for the second
file.
Similarly, Redo operator can also be explored to move back in loop.
Searching patterns in 2 data files:
In bioinformatics work, it is often necessary to search a list of names from a file from another file, that either has a list of sequences or blast output or anything else. So, the trick to reduce the search time will be keep the larger file as the outer loop(mostly as a while loop), the inside loop could be a while loop or a for loop, depending on the file size. If there is a for loop for the inner loop, then it is possible to get rid of the element that has just had a match to the outer element. This way with every iteration, the inner loop gets shorter and the time spent in searching gets shorter. In perl it is quite easy to remove an array element by simply using a delete function.
delete($array[index]) will do the job. But just delete removes the element while leaving $array[index] space empty. In order to remove the element along with index use splice function instead.
splice(@array,index,offset) --> will remove offset number of elements and indices starting from index. i.e; splice(@array,2,7) will remove 7 indices starting from 2.
The whole construct may look something like this:
while(Passing Array by Reference){ if(/Query= (\S+)/){ $flag = 0; for(my $i; $i<=$#arr;$i++){ my @tmp = split(/\t/,$arr[$i]); if($1 eq $tmp[0]){ print "$_"; $flag = 1; delete($arr[$i]); # or splice(@arr,$i,1); last; } } next; } if($flag){ print "$_"; } }
If you have a subroutine and you need to pass more than one array, then don't pass by value. If passed by value then the list is flattened and the first and second element of the array is passed as 2 arrays. Example: @arr1 = [1,2,3];
@arr2 = [6,7,8];
func(@arr1,@arr2);
will print: Array1 is 1 and array2 is 2
sub func
{
my @a = shift;
my @b = shift;
print "Array1 is @a and array2 is @b\n";
}
In order to get the values intact pass by reference:
func(\@arr1,\@arr2);
in
sub func{
$a = shift;
$b = shift;
@arr1 = @$a; # de-referencing array
@arr2 = @$b;
Here is a time and space saver if you want to sort a hash by value and iterate over the sorted list:
# sorting tmp hash by value
foreach my $sorted(sort {$tmp{$b} <=> $tmp{$a}} keys %tmp){
my %geneHash;
for my $gene(keys %{$HOH1{$sorted}}){
my @coord = sort{$a <=> $b} @{ $HOH1{$sorted}{$gene}{'pos'}};
my $strand1 = $HOH1{$sorted}{$gene}{'strand'};
$geneHash{$gene} = $coord[0];
}
foreach my $sortedgene(sort {$geneHash{$a} <=> $geneHash{$b}} keys %geneHash ){
print FH "$sortedgene\t$serielNumber\n";
print FH1 ">
$serielNumber\n";
print FH1 "$transcript{$sortedgene}\n";
`sed 's/\t$sortedgene/$serielNumber/' output.gff`;
$serielNumber++;
}
}
# One variation if you want to sort the sequence file in descending order of their length. step -1:
Read fasta file into a hash called as %hash
# sort hash by the length of the value
foreach my $scaffold(sort {length($hash{$b}) <=> length($hash{$a}) } keys %hash) {
print ">scaffold_$num\n$hash{$scaffold}\n";
print FH "$scaffold\tscaffold_$num\t".length($hash{$scaffold})."\n"; $num++;
}
To make it to the ascending order simply change this to:
foreach my $scaffold(sort {length($hash{$a}) <=> length($hash{$b}) } keys %hash)
Thursday, February 12, 2009
Finding overlap
In sequence analysis, it is a common practice to find overlaps between 2 sets of features. This may be a coding sequence with repeats or coding sequences from 2 different prediction programs or 2 different coding sequences and so on. If data is present in gff files and are read into hashes with gene being the key holding the positions in an array, then the comparison can be made the following ways:
Lets find overlap between a gene and a repeat element.
geneLeft, repeatLeft be the first element of gene and repeat array respectively and geneRight, repeatRight be the last element of the gene and repeat array respectively.
#if repeat elements are occurring towards the left side of the gene element:
# and overlaps at the left hand side
repeatLeft <= geneLeft and repeatRight >= geneLeft # Note here Not repeatRight <= geneRight -> This condition will be true even if the repeat is occuring way upstream of the gene element and even if they don't overlap
# If repeat occurs at the right han side of the gene and overlaps at the right side
repeatLeft <= geneRight and repeatRight >= geneRight
# If repeat element lies within the gene element
geneLeft <= repeatLeft and repeatRight <= geneRight
#If gene element occurs inside the repeat element
repeatLeft <= geneLeft and geneRight <= repeatRight
If any of these conditions become true then it will find overlap(Join them with 'or')
if( ($coord1[$i] <= $coord[0] && $coord1[$i+1] >= $coord[-1]) ||
($coord1[$i] >= $coord[0] && $coord1[$i+1] <= $coord[-1]) ||
($coord1[$i] <= $coord[0] && $coord1[$i+1] >= $coord[0]) ||
($coord1[$i] >= $coord[0] && $coord1[$i] <= $coord[-1] )){
Tuesday, February 10, 2009
Using multidimensional hashes in storing sequence objects
One of the most important functions of multidimensional perl hashes has been their usage in reading sequence related data. The most common data type one reads from a genome sequencing prospective is a gff file. A gff file has the following attributes that are most likely captured in a data structure:
1. Name of the scaffold or chromosome on which the gene resides.
2. Name of the gene
3. Gene positions
4. Strand
Other attributes are less important, so lets ignore them at the moment.
So, now a scaffold will have multiple genes and a gene will have multiple locations. The ideal data structure that can store all these information will be a has of hashes and one of the hash holds arrays. In other words:
scaffold_1 => {
gene1 => { pos => [positions], strand => + or minus},
gene2 => { pos => [positions], strand => + or minus},
.......
},
scaffold_2 => {
gene1 => {pos => [positions], strand => + or minus},
gene2 => {pos => [positions], strand => + or minus},
.......
},
.....
Populating this hash from gff file:
sub test_gff{
my $fileName = shift;
my %HOH;
open PRED, $fileName or die "Can't open file $!\n";
while(){
chomp;
my @line = split(/\t/,$_);
push(@{$HOH{$line[0]}->{$line[8]}->{'pos'}}, $line[3], $line[4]);
$HOH{$line[0]}->{line[8]}->{'strand'}, $line[5];
}
close PRED;
return %HOH;
}
Now Printing the values:
my %HOH1 = &utility::test_gff($ARGV[0]); # Read the file
for my $scaffold(keys %HOH1){
for my $gene(keys %{$HOH1{$scaffold}}){
my @coord = @{ $HOH1{$scaffold}{$gene}{'pos'}};
my $strand = $HOH1{$scaffold}{$gene}{'strand'};
print "$scaffold -> $gene -> @coord -> $strand\n";
}
}
# If you want to sort the co-ordinates,Then that can be done as below:
my @coord = sort{$a <=> $b} @{ $HOH1{$scaffold}{$gene}{'pos'}};
# Values will be printed as :
Contig632 -> 713969 -> 1687 3391 -> +
Contig1122 -> 707832 -> 3109 3183 3194 3200 3200 3232 3232 3251 -> +
Contig1294 -> 714344 -> 3744 4473 -> +
Contig1294 -> 714343 -> 24 1652 -> -
Contig772 -> 707541 -> 14 64 126 147 147 182 -> +
Contig772 -> 713832 -> 4 81 126 147 147 194 -> +
Contig772 -> 711369 -> 1 64 130 147 147 204 -> +
Cool...
Adding one more dimension:
Mostly in draft genome sequences the genome assembly is not in great shape, so most often same genes can be found in multiple locations(Not necessarily through gene duplication). Sometimes the same gene could be present in the same scaffold in more than one location. In that case to stop over writing or appending the positions for the same gene, another check can be implemented, that is the gene serial number. So, you can just add position as another attribute with seriel number being the key.
push(@{$HOH{$line[0]}->{$line[8]}->{$serialNum}->{'pos'}}, $line[3], $line[4]);
$HOH{$line[0]}->{line[8]}->{$serialNum}->{'strand'}, $line[5];
And while iterating add one more for loop:
for my $scaffold(keys %HOH1){
for my $gene(keys %{$HOH1{$scaffold}}){
for my $serialNum(keys %{%HOH1{$scaffold}{$gene}}){
my @coord = @{ $HOH1{$scaffold}{$gene}{$serialNum}{'pos'}};
my $strand = $HOH1{$scaffold}{$gene}{$serialNum}{'strand'};
print "$scaffold -> $gene -> @coord -> $strand\n";
}
}
The ease at which we can use perl data structures make it so desirable. The coder does not need to worry about memory allocation stuff. Earlier I used to use linked list in C to store sequence data. Then I will be stuck in debugging stuff a lot. Now after I discovered this, I have almost stopped coding in C. Happy perl coding..
Thursday, January 29, 2009
Editing publication quality images with GIMP
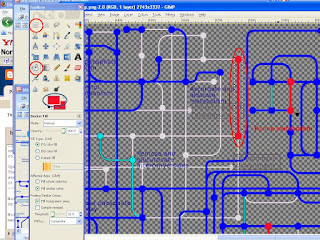
GIMP stand for GNU Image Manipulating Program. I have this software in my computer on and off for the last few years. I was adamant to not budge for paid softwares like Adobe photo shop and like. Gimp used to sit there in my computer and annoy me with its small icon, so I used to uninstall it often. When I needed to manipulate some figures for publication, I would install it back on. Not until very recently I had no clue how powerful and useful gimp can be. It can even beat the best image editing softwares in market..
Challenges:
Recently, I faced a challenge merging 3 pathway maps. They can be as complicated as a circuit diagram!! I had to basically merge Image 1 and Image 2 and make a composite image12. Then I had to merge this with image 3 in such a way that, imae3 - image12 will be in a different color and image12 - image3 will be in a different color.
Open image 1 as layer(file --> open as layer), then open image 2 as layer as well. Then go to layer menu --> Merge down --> save image as image12. In image12, you will see the extra portions of image 1 as little lighter objects, but image 2 appears as it is. Now open image12 as layer then open image 3 as layer. Then go to layer menu --> merge down --> save as image123.
Now in image123 you see the extra portions in 12 that is not present in 3 as lighter objects. Well enough..
Now do the reverse. Open image 3 as layer first followed by image 12 as layer. Then merge as before and name it as image 312.
What you will see is in image 312 --> All that is present extra in 3 are not present in image 12 will appear as solid bordered transparent object. Distinct from the upper layer. In Image 123 you will see just the reverse. The ones that are present extra in image 12 and are not present in image 3. Now after all these, you can happily open image 312 and start playing around with colors. Open image 312 and as shown in figure select the shape from the toolbox menu. Select the area neatly that need to be colored differently. Then choose color. Then select color menu and just click on the selected area in the image. Viola.. that changes the color so well....
Friday, January 16, 2009
The power of vim
vim is a very powerful text editor having many great features that can prove to be a time saver if you are editing a large file.
I use the substitution operation in vim more often than not. One can a do a number of things with just very little practice.
1. Make a newline:
%s/$/\r/g # will make the end of the line into a new line character.
2. Replace annoying ^M characters that often happens from a DOS to unix transition
%s/ctrlVM/BY anything you like/ # Remember here ctrl-V makes a ^ and M makes an M
3. If you want to substitute something from line 20,30 here is how you can do
:20,30s/old/new/ -> remember here no '%' before 's'
4. If you want to change the order of two words from Hypothetical protein -> protein Hypothetical
:%s/\(Hypothetical\) \(protein\)/\2 \1/ -> This will change all occurrences of Hypothetical protein to protein Hypothetical.
Some More Tricks in vim
Delete lines that contain pattern:
:g/pattern/d
Delete all empty lines:
:g/^$/d
Delete lines in range that contain pattern:
:20,30/pattern/d
or with marks a and b:
:'a,'b/pattern/d
Substitute all lines for first occurance of pattern:
:%s/pattern/new/
:1,$s/pattern/new/
Substitute all lines for pattern globally (more than once on the line):
:%s/pattern/new/g
:1,$s/pattern/new/g
Find all lines containing pattern and then append -new to the end of each line:
:%s/\(.*pattern.*\)/\1-new/g
Substitute range:
:20,30s/pattern/new/g
with marks a and b:
:'a,'bs/pattern/new/g
Swap two patterns on a line:
:s/\(pattern1\)\(pattern2\)/\2\1/
Capitalize the first lowercase character on a line:
:%s/\([a-z]\)/\u\1/
more concisely:
:%s/[a-z]/\u&/
Capitalize all lowercase characters on a line:
:%s/\([a-z]\)/\u\1/g
more concisely:
:s/[a-z]/\u&/g
Capitalize all characters on a line:
:s/\(.*\)/\U\1\E/
Example: In a sequence file, if you have multiple headers and you are particularly
interested in substituting line Hp_ENSC_10a23 -> Hp_ENSC_10A23 without touching any
other line this is for you..
:%s/ENSC_\(.*\)\([a-z]\)/ENSC_\1\u\2/g
Regular Expressions:
Regular expressions in unix is more or less similar like perl - ya perl borrowed regexp from Unix..
But using substitution, one need to take care of adding '\' (esc) character in order to match a value.
For example if a line with super_2[TAB]5997239[TAB]5997668 needs to change to super_2:5997239-5997668, then instead of using '\S+' as the first non space patter use \(\S\+\). So, in other words the syntax will be
:%s/^\(\S\+\)\t\(\S\+\)\t/\1:\2-/g
Capitalize the first character of all words on a line:
:s/\<[a-z]/\u&/g
Uncapitalize the first character of all words on a line:
:s/\<[A-Z]/\l&/g
Change case of character under cursor:
~
Change case of all characters on line:
g~~
Change case of remaining word from cursor:
g~w
Increment the number under the cursor:
Decrement the number under the cursor:
redraw:
Turn on line numbering:
:set nu
Turn it off:
:set nonu
Number lines (filter the file through a unix command and replace with output):
:%!cat -n
Sort lines:
:%!sort
Sort and uniq:
:%!sort -u
Read output of command into buffer:
:r !ls -l
Refresh file from version on disk:
:e!
Open a new window:
n
Open a new window with the same file (split):
s
Split window vertically:
v
Close current window:
c
:q
Make current window the only window:
o
Cycle to next window:
w
Move to window below current window:
j
Move to window above current window:
k
Move to window left of current window:
h
Move to window right of current window:
l
Set textwidth for automatic line-wrapping as you type:
:set textwidth=80
Turn on syntax highlighting
:syn on
Turn it off:
:syn off
Force the filetype for syntax highlighting:
:set filetype=python
:set filetype=c
:set filetype=php
Use lighter coloring scheme for a dark background:
:set background=dark
Htmlize a file using the current syntax highlighting:
:so $VIMRUNTIME/syntax/2html.vim
Or, htmlize from a command prompt:
in 2html.sh put:
#!/bin/sh
vim -n -c ':so $VIMRUNTIME/syntax/2html.vim' -c ':wqa' $1 > /dev/null 2> /dev/null
Now just run: shell> 2html.sh foo.py
Getting rid of tab while copy pasting from a vim
edited file:
By default when you try to copy paste a block from a file edited with vim, it automatically appends character in the beginning of each line. In order to get rid of that in your .vimrc file put a line saying "set paste". This will do the job.
File Recovery:
Many times while editing a file the connection to the server may be lost. This is what I face all the time. In that case you can recover all the changes you made earlier by saying vim -r .file_name.swp
Executing Shell Commands
You can execute a command with the shell, for example, you can compile you C code when editing it, :!gcc kernel.c, or :!gcc % to compile the current editing file. Executing other commands is the same.
Keyword Matching
If you are lazy as me, you will find this function wonderful :) In insert mode, you can type a few characters of a word, e.g., if there is a variable called phosphatidylinositolphosphate, you may just type pho then C-p. Vim will find out the last word in the file started with characters pho, if it is not you want, you can re-type C-p to match the further previous ones. Similarly, C-n can do that for finding the next matches. Therefore you do not worry about the mistyping of the long variables, or uncommon-used words.
Insert/Delete an indent
C-T/C-D insert/delete an indent in the current line, no matter what column the cursor is in. There are handy hot keys when you are editing language with ambiguous block boundary such as Python, which you have to delete the indent yourself when necessary.
Settings for .vimrc
Some useful setting and mapping for .vimrc are listed as follows:
set nocp " nocompatible
set sw=4 " shiftwidth
set et " expandtab
set wm=8 " wrapmargin
set bs=2 " backspace
set ru " ruler
set ic " ignorecase
set is " incsearch
set scs " smartcase: override the 'ic' when searching
" if search pattern contains uppercase char
set vb t_vb= " set visual bell and disable screen flash
set backup
NOTE
Sometimes find replace and delete lines with a particular pattern can very quickly done using sed. Yes, vim takes longer time to complete a request if the file is large. So, instead try sed.
sed '/^--/d' lane7.1 > tmp # Will delete all lines with pattern --
# from file lane7.1 and will print in tmp
# -- comes as a pattern delimiter from 'grep'
sed 's/^@/>/g' lane7.1 > tmp # Will replace all lines beginning with @ to >
# This is useful in converting fastq files to fasta
I use the substitution operation in vim more often than not. One can a do a number of things with just very little practice.
1. Make a newline:
%s/$/\r/g # will make the end of the line into a new line character.
2. Replace annoying ^M characters that often happens from a DOS to unix transition
%s/ctrlVM/BY anything you like/ # Remember here ctrl-V makes a ^ and M makes an M
3. If you want to substitute something from line 20,30 here is how you can do
:20,30s/old/new/ -> remember here no '%' before 's'
4. If you want to change the order of two words from Hypothetical protein -> protein Hypothetical
:%s/\(Hypothetical\) \(protein\)/\2 \1/ -> This will change all occurrences of Hypothetical protein to protein Hypothetical.
Some More Tricks in vim
Delete lines that contain pattern:
:g/pattern/d
Delete all empty lines:
:g/^$/d
Delete lines in range that contain pattern:
:20,30/pattern/d
or with marks a and b:
:'a,'b/pattern/d
Substitute all lines for first occurance of pattern:
:%s/pattern/new/
:1,$s/pattern/new/
Substitute all lines for pattern globally (more than once on the line):
:%s/pattern/new/g
:1,$s/pattern/new/g
Find all lines containing pattern and then append -new to the end of each line:
:%s/\(.*pattern.*\)/\1-new/g
Substitute range:
:20,30s/pattern/new/g
with marks a and b:
:'a,'bs/pattern/new/g
Swap two patterns on a line:
:s/\(pattern1\)\(pattern2\)/\2\1/
Capitalize the first lowercase character on a line:
:%s/\([a-z]\)/\u\1/
more concisely:
:%s/[a-z]/\u&/
Capitalize all lowercase characters on a line:
:%s/\([a-z]\)/\u\1/g
more concisely:
:s/[a-z]/\u&/g
Capitalize all characters on a line:
:s/\(.*\)/\U\1\E/
Example: In a sequence file, if you have multiple headers and you are particularly
interested in substituting line Hp_ENSC_10a23 -> Hp_ENSC_10A23 without touching any
other line this is for you..
:%s/ENSC_\(.*\)\([a-z]\)/ENSC_\1\u\2/g
Regular Expressions:
Regular expressions in unix is more or less similar like perl - ya perl borrowed regexp from Unix..
But using substitution, one need to take care of adding '\' (esc) character in order to match a value.
For example if a line with super_2[TAB]5997239[TAB]5997668 needs to change to super_2:5997239-5997668, then instead of using '\S+' as the first non space patter use \(\S\+\). So, in other words the syntax will be
:%s/^\(\S\+\)\t\(\S\+\)\t/\1:\2-/g
Capitalize the first character of all words on a line:
:s/\<[a-z]/\u&/g
Uncapitalize the first character of all words on a line:
:s/\<[A-Z]/\l&/g
Change case of character under cursor:
~
Change case of all characters on line:
g~~
Change case of remaining word from cursor:
g~w
Increment the number under the cursor:
Decrement the number under the cursor:
redraw:
Turn on line numbering:
:set nu
Turn it off:
:set nonu
Number lines (filter the file through a unix command and replace with output):
:%!cat -n
Sort lines:
:%!sort
Sort and uniq:
:%!sort -u
Read output of command into buffer:
:r !ls -l
Refresh file from version on disk:
:e!
Open a new window:
Open a new window with the same file (split):
Split window vertically:
Close current window:
:q
Make current window the only window:
Cycle to next window:
Move to window below current window:
Move to window above current window:
Move to window left of current window:
Move to window right of current window:
Set textwidth for automatic line-wrapping as you type:
:set textwidth=80
Turn on syntax highlighting
:syn on
Turn it off:
:syn off
Force the filetype for syntax highlighting:
:set filetype=python
:set filetype=c
:set filetype=php
Use lighter coloring scheme for a dark background:
:set background=dark
Htmlize a file using the current syntax highlighting:
:so $VIMRUNTIME/syntax/2html.vim
Or, htmlize from a command prompt:
in 2html.sh put:
#!/bin/sh
vim -n -c ':so $VIMRUNTIME/syntax/2html.vim' -c ':wqa' $1 > /dev/null 2> /dev/null
Now just run: shell> 2html.sh foo.py
Getting rid of tab while copy pasting from a vim
edited file:
By default when you try to copy paste a block from a file edited with vim, it automatically appends
File Recovery:
Many times while editing a file the connection to the server may be lost. This is what I face all the time. In that case you can recover all the changes you made earlier by saying vim -r .file_name.swp
Executing Shell Commands
You can execute a command with the shell, for example, you can compile you C code when editing it, :!gcc kernel.c, or :!gcc % to compile the current editing file. Executing other commands is the same.
Keyword Matching
If you are lazy as me, you will find this function wonderful :) In insert mode, you can type a few characters of a word, e.g., if there is a variable called phosphatidylinositolphosphate, you may just type pho then C-p. Vim will find out the last word in the file started with characters pho, if it is not you want, you can re-type C-p to match the further previous ones. Similarly, C-n can do that for finding the next matches. Therefore you do not worry about the mistyping of the long variables, or uncommon-used words.
Insert/Delete an indent
C-T/C-D insert/delete an indent in the current line, no matter what column the cursor is in. There are handy hot keys when you are editing language with ambiguous block boundary such as Python, which you have to delete the indent yourself when necessary.
Settings for .vimrc
Some useful setting and mapping for .vimrc are listed as follows:
set nocp " nocompatible
set sw=4 " shiftwidth
set et " expandtab
set wm=8 " wrapmargin
set bs=2 " backspace
set ru " ruler
set ic " ignorecase
set is " incsearch
set scs " smartcase: override the 'ic' when searching
" if search pattern contains uppercase char
set vb t_vb= " set visual bell and disable screen flash
set backup
NOTE
Sometimes find replace and delete lines with a particular pattern can very quickly done using sed. Yes, vim takes longer time to complete a request if the file is large. So, instead try sed.
sed '/^--/d' lane7.1 > tmp # Will delete all lines with pattern --
# from file lane7.1 and will print in tmp
# -- comes as a pattern delimiter from 'grep'
sed 's/^@/>/g' lane7.1 > tmp # Will replace all lines beginning with @ to >
# This is useful in converting fastq files to fasta
Wednesday, January 14, 2009
Plotting Histograms and boxplot with R
Plotting huge files in R can be a cake walk. Here is how you can proceed:
Suppose say you have a huge file with 2 million entries in[TAB] format.
Then go ahead and read the file in the following format:
>data <- read.table(file="fileName",head=TRUE,sep=",")
# read.table is a file reading function
# Note here, if you have no header then say head=FALSE and if you data is separated by
# \t then say sep="\t"
#You may like to see summary of your data by saying
> summary(data)
V1 V2
HPAA-aaa57a02.b1: 1 Min. :0.000000
HPAA-aaa57a02.g1: 1 1st Qu.:0.009985
HPAA-aaa57a03.b1: 1 Median :0.020565
HPAA-aaa57a03.g1: 1 Mean :0.021674
HPAA-aaa57a04.b1: 1 3rd Qu.:0.033898
HPAA-aaa57a04.g1: 1 Max. :0.065789
(Other) :1140845
# Pretty Cool..
If you have not written down the column names in the data file, don't worry R provides one for you.
>names(data)
>[1] "V1" "V2"
# So, V1 and V2 are your column names.
# To Plot a Histogram simply say
>hist(data$V2)
# If you did not select any output device by default it will plot as a Rplots.ps file
# on your working directory
# You can however copy this to a file of your interest
>dev.copy(postscript,"myfile.ps",height=7,width=7)
# Here Height and width are in inches
# Sometimes it does not allow you to do a dev.copy when your dev.list() is NULL.
# You can populate it by first plotting something or by doing a hist(data$V2)
# or just a plot(data$V2)
# Then do
>hist(data$V2)
# You can check how many devices are attached to your screen by doing a
> dev.list()
postscript postscript postscript
2 3 4
# You can switch off your current device by doing a
>dev.off()
postscript
2
> dev.list()
postscript postscript
2 3
# I Plotted my histogram using the following command:
5
> hist(data$V2,breaks=100,main="Trace File Blast hit Frequency plot, cutoff=30"
,xlab="Y=M/(R+L - 2O)",col="red",las=1,freq=TRUE)
> lines(density(data$V2))
This will save the file in the current file "myfile.ps". Every subsequent call to a plot function will keep plotting in different pages of the same file.
Alternatively a truehist() can be called to plot histograms. But before that do a
>require(MASS)
> truehist(data$V2,nbins=100,main="Trace File Blast hit Frequency plot, cutoff=
30",xlab="Y=M/(R+L - 2O)",col="blue",las=1,prob=TRUE)
Making a boxplot:
Plotting a boxplot is equally easier using R. Follow the following commands:
data <- read.table(file="bgcorrect.out",head=FALSE,sep="\t")
summary(data)
#boxplot(data$vals,main='normalized data after synthetic normalization', ylab='G
ene Expression values');
boxplot(log2(data))
help(boxplot)
SAVING OUTPUT AS TEXT FILE
#In case you are just interested to save the histogram output file into a text file
> h <- hist(data$V2,breaks=100,plot=FALSE) # Plot= FALSE will not plot, default true
> save(h,ascii=TRUE,file="data.txt") # Will save data in ascii format, default=FALSE
# If you want to read the content of h
>h
# You will get the following
# $breaks -> break points
# $count -> frequency
# $density -> plot density
# $intensities -> plot intensity
# $mids -> mid points of data
So, just printing a single variable will be
>h$mids # Will print the mid point..
Suppose say you have a huge file with 2 million entries in
Then go ahead and read the file in the following format:
>data <- read.table(file="fileName",head=TRUE,sep=",")
# read.table is a file reading function
# Note here, if you have no header then say head=FALSE and if you data is separated by
# \t then say sep="\t"
#You may like to see summary of your data by saying
> summary(data)
V1 V2
HPAA-aaa57a02.b1: 1 Min. :0.000000
HPAA-aaa57a02.g1: 1 1st Qu.:0.009985
HPAA-aaa57a03.b1: 1 Median :0.020565
HPAA-aaa57a03.g1: 1 Mean :0.021674
HPAA-aaa57a04.b1: 1 3rd Qu.:0.033898
HPAA-aaa57a04.g1: 1 Max. :0.065789
(Other) :1140845
# Pretty Cool..
If you have not written down the column names in the data file, don't worry R provides one for you.
>names(data)
>[1] "V1" "V2"
# So, V1 and V2 are your column names.
# To Plot a Histogram simply say
>hist(data$V2)
# If you did not select any output device by default it will plot as a Rplots.ps file
# on your working directory
# You can however copy this to a file of your interest
>dev.copy(postscript,"myfile.ps",height=7,width=7)
# Here Height and width are in inches
# Sometimes it does not allow you to do a dev.copy when your dev.list() is NULL.
# You can populate it by first plotting something or by doing a hist(data$V2)
# or just a plot(data$V2)
# Then do
>hist(data$V2)
# You can check how many devices are attached to your screen by doing a
> dev.list()
postscript postscript postscript
2 3 4
# You can switch off your current device by doing a
>dev.off()
postscript
2
> dev.list()
postscript postscript
2 3
# I Plotted my histogram using the following command:
5
> hist(data$V2,breaks=100,main="Trace File Blast hit Frequency plot, cutoff=30"
,xlab="Y=M/(R+L - 2O)",col="red",las=1,freq=TRUE)
> lines(density(data$V2))
This will save the file in the current file "myfile.ps". Every subsequent call to a plot function will keep plotting in different pages of the same file.
Alternatively a truehist() can be called to plot histograms. But before that do a
>require(MASS)
> truehist(data$V2,nbins=100,main="Trace File Blast hit Frequency plot, cutoff=
30",xlab="Y=M/(R+L - 2O)",col="blue",las=1,prob=TRUE)
summary(data)
#boxplot(data$vals,main='normalized data after synthetic normalization', ylab='G
ene Expression values');
boxplot(log2(data))
help(boxplot)
SAVING OUTPUT AS TEXT FILE
#In case you are just interested to save the histogram output file into a text file
> h <- hist(data$V2,breaks=100,plot=FALSE) # Plot= FALSE will not plot, default true
> save(h,ascii=TRUE,file="data.txt") # Will save data in ascii format, default=FALSE
# If you want to read the content of h
>h
# You will get the following
# $breaks -> break points
# $count -> frequency
# $density -> plot density
# $intensities -> plot intensity
# $mids -> mid points of data
So, just printing a single variable will be
>h$mids # Will print the mid point..
Friday, January 09, 2009
RepeatModeler Woes...
Continued from my last post..
Folks, if you are tempted to run a de novo repeat finder such as RepeatModeler, then this is for you.
I happened to get copies of all the dependencies along with RepeatModeler and got them installed in my linux machine. Here is what is not documented in any installation manual.
Installation Caveats:
1. If you installed RepeatMasker without getting a copy of RepBase, it will not complain and installation will be fine. But when you complete installation of RepeatModeler(If you could), then it will complain big time - "you did not install RepBase on repeatMasker", so go back and re-configure RepeatMasker all over again.
SO, LESSON NO 1 ALWAYS GET REPBASE BEFORE INSTALLING REPEATMASKER
2. TRFinder is named as trfLinux.. when you obtain it from the download site. The installation program looks for just "trf" program in the folder. So, RENAME the executable to trf before installation.
3. The biggest trouble comes with installation of RepeatModeler. It will run smooth till it asks you the path for wublast installation. WUBLAST is currently named as ABBLast and is no longer available. You only get the 1998 version from the web site. If you are lucky and you have a copy of wublast, then the installation complains about xdformat.
ERROR:
/home/sutripa/wublast/xdformat is too old. Must have one dated 3/16/2005 or new
er. Install a newer version of wublast and re-run configure.
In order to fix this, you can do little bit of hacking:
Go to the "configure" file in repeatmodeler package and comment the block
#unless ( $result =~ /option requires an argument -- m/ ) {
# die "$wuLocation/xdformat is too old. Must have one dated 3/16
/2005 "
# . "or newer. Install a newer version of wublast and re-run
"
# . "configure.\n";
#}
(This is located between line number 247 - 255)
4. While giving the path for RECON, say $RECON_PATH/bin. Otherwise configuration script will not find the executables.
Once all these are taken care of installation of repeatmodeler can be done.
RUNNING REPEATMODELER:
Again this is not any lesser challenge than installation.
Goto RepeatModeler file and on line 234 do the following:
Replace line
#my $dbIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $genomeDB`;
with
my $dbIndex = `$RepModelConfig::XDFORMAT_PRGM -n -r $genomeDB`;
AND ALSO Line number 500
Replace line
#my $sampleIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $sampleFa
staFile`;
with
my $sampleIndex = `$RepModelConfig::XDFORMAT_PRGM -n -r $sampleFastaF
ile`;
Use the README file for creating a database using the xdformat program from wublast.
/BuildXDFDatabase -name elephant elephant.fa
IMPORTANT:
[Before Running this, check your fasta file. Make the header of fasta as >string[SPACE]number format, where the number could ideally be the length of the sequence. Leave nothing else trailing on the header.]
If you fail to do this, while running wublast the program is going to complain
either of these Two. Either sequence not found OR Database is empty.
After doing all this, you may like to run the program as
RepeatModeler -database Ha.test >& Ha.out
Alas!!You thought the problem is finally solved and go check the run file. It still has error like:
FastaDB:_cleanIndexAndCompact - ERROR: Multiple fasta seqs appear on one line (>
seq-3_1-40000) and possibly more! Ignoring all entries on this line.
FastaDB:_cleanIndexAndCompact - ERROR: Multiple fasta seqs appear on one line (>
seq-1_1280001-1320000) and possibly more! Ignoring all entries on this line.
FastaDB:_cleanIndexAndCompact - ERROR: Multiple fasta seqs appear on one line (>
seq-1_520001-560000) and possibly more! Ignoring all entries on this line.
This is beacuse there is a silly bug in line 1590.
Go check one of the directories RM_12849.TueJan131331092009/round-1
you will see the header is printed like this: >>seq-3_1-40000
Now go to line 1590 in RepeatModeler and Replace
#print FASTA ">$seqID" . "_$start-$end\n";
With
print FASTA "$seqID" . "_$start-$end\n";
Now you can run the program.
Even after doing all these, I still have no luck in running this script. The run file ended up giving me a "unix broken pipe" error which is way beyond my control to fix.
Argh...
Folks, if you are tempted to run a de novo repeat finder such as RepeatModeler, then this is for you.
I happened to get copies of all the dependencies along with RepeatModeler and got them installed in my linux machine. Here is what is not documented in any installation manual.
Installation Caveats:
1. If you installed RepeatMasker without getting a copy of RepBase, it will not complain and installation will be fine. But when you complete installation of RepeatModeler(If you could), then it will complain big time - "you did not install RepBase on repeatMasker", so go back and re-configure RepeatMasker all over again.
SO, LESSON NO 1 ALWAYS GET REPBASE BEFORE INSTALLING REPEATMASKER
2. TRFinder is named as trfLinux.. when you obtain it from the download site. The installation program looks for just "trf" program in the folder. So, RENAME the executable to trf before installation.
3. The biggest trouble comes with installation of RepeatModeler. It will run smooth till it asks you the path for wublast installation. WUBLAST is currently named as ABBLast and is no longer available. You only get the 1998 version from the web site. If you are lucky and you have a copy of wublast, then the installation complains about xdformat.
ERROR:
/home/sutripa/wublast/xdformat is too old. Must have one dated 3/16/2005 or new
er. Install a newer version of wublast and re-run configure.
In order to fix this, you can do little bit of hacking:
Go to the "configure" file in repeatmodeler package and comment the block
#unless ( $result =~ /option requires an argument -- m/ ) {
# die "$wuLocation/xdformat is too old. Must have one dated 3/16
/2005 "
# . "or newer. Install a newer version of wublast and re-run
"
# . "configure.\n";
#}
(This is located between line number 247 - 255)
4. While giving the path for RECON, say $RECON_PATH/bin. Otherwise configuration script will not find the executables.
Once all these are taken care of installation of repeatmodeler can be done.
RUNNING REPEATMODELER:
Again this is not any lesser challenge than installation.
Goto RepeatModeler file and on line 234 do the following:
Replace line
#my $dbIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $genomeDB`;
with
my $dbIndex = `$RepModelConfig::XDFORMAT_PRGM -n -r $genomeDB`;
AND ALSO Line number 500
Replace line
#my $sampleIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $sampleFa
staFile`;
with
my $sampleIndex = `$RepModelConfig::XDFORMAT_PRGM -n -r $sampleFastaF
ile`;
Use the README file for creating a database using the xdformat program from wublast.
IMPORTANT:
[Before Running this, check your fasta file. Make the header of fasta as >string[SPACE]number format, where the number could ideally be the length of the sequence. Leave nothing else trailing on the header.]
If you fail to do this, while running wublast the program is going to complain
either of these Two. Either sequence not found OR Database is empty.
After doing all this, you may like to run the program as
RepeatModeler -database Ha.test >& Ha.out
Alas!!You thought the problem is finally solved and go check the run file. It still has error like:
FastaDB:_cleanIndexAndCompact - ERROR: Multiple fasta seqs appear on one line (>
seq-3_1-40000) and possibly more! Ignoring all entries on this line.
FastaDB:_cleanIndexAndCompact - ERROR: Multiple fasta seqs appear on one line (>
seq-1_1280001-1320000) and possibly more! Ignoring all entries on this line.
FastaDB:_cleanIndexAndCompact - ERROR: Multiple fasta seqs appear on one line (>
seq-1_520001-560000) and possibly more! Ignoring all entries on this line.
This is beacuse there is a silly bug in line 1590.
Go check one of the directories RM_12849.TueJan131331092009/round-1
you will see the header is printed like this: >>seq-3_1-40000
Now go to line 1590 in RepeatModeler and Replace
#print FASTA ">$seqID" . "_$start-$end\n";
With
print FASTA "$seqID" . "_$start-$end\n";
Now you can run the program.
Even after doing all these, I still have no luck in running this script. The run file ended up giving me a "unix broken pipe" error which is way beyond my control to fix.
Argh...
RepeatMasker blues..
I read rave reviews on RepeatMasker for repeat finding, so decided to get hold of a copy and get it used. I used this software before and did not like it much, but after getting some leading genome research institutes using it for their newly sequenced genomes, I got tempted. I used to use TRFinder from Boston university for finding direct repeats earlier.
I went to the web page of RepeatMasker that directs me to an array of other dependencies to be installed first. RepeatMasker depends on a repeat library RepBase which can be obtained from http://www.girinst.org. You go there and have to get yourself registered. Your registration need to be accepted by the group and then they send you an email carrying your login id and password. This may take upto 2 working days. Only after this you can download the RepBase database.
However, the trouble with RepBase is, it is a predefined library and will not detect any new repeat that may be there in your genome. So, the best option is to run a "De Novo" repeat finder. From the same web site you can get a link to RepeatModeler that acts as a wrapper around some de novo repeat finders such as RepeatScout and RECON.
Fair enough, then you go to install RepeatModeler...
This web page indicates you have to have a number of dependencies:
-- Of course you need to have perl 5.8 or above installed first.
-- you will need the Trfinder that I was mentioning earlier.
-- then you will need to download and install repeatmasker. This will not install unless you have RepBase in place.
--For RepeatMasker, you will also need phred/phrap/cross_match package. This can be done by writing to the authors and getting a copy by email. This may take a few business days.
--After you are done with repeatmasker you will need RECON to be installed
--Then RepeatScout need to be downloaded and installed.
--You will need WUBLAST also to be installed. The recommended site from the repeatmodeler page points to WUBLAST site at http://blast.wustl.edu/. When you go there you get redirected to http://www.advbiocomp.com . In advbiocomp site, they say that wublast is no longer supported and you can get an older version of WUblast for free. Then you get the binaries, inflate and keep them in place.
Now the final part: Installation of RepeatModeler. You do a ./configure. It asks you perl path, repeatMasker path etc. When it comes to WuBlast path, it exits saying that you xdformat program is older than 2005. What the hell, you back and check in wublast site, that says the program is as old as 1998. There are no newer version. There you get stuck.
Out of frustartion, I went and looked at the configure file in RepeatModeler. In line 237 there is a statement:
else {
my $result = `$wuLocation/xdformat -m 2>&1`;
# Two responses are expected:
# Good Version:
# /blah/wublast/xdformat: option requires an argument -- m
# Old Version:
# /blah/wublast/xdformat: invalid option -- m
unless ( $result =~ /option requires an argument -- m/ ) {
die "$wuLocation/xdformat is too old. Must have one dated 3/16/
5 "
. "or newer. Install a newer version of wublast and re-run
. "configure.\n";
}
I went back and asked the authors of xdformat, what this -m option means, they said to ignore it. Now I can change this, if only I find what is the substitute of this. Because my search in repeatModeler directory says me -m2 option is used:
gus@tyler-lab:~/RepeatModeler$ grep --regex=-m -r .
./configure: my $result = `$wuLocation/xdformat -m 2>&1`;
./Refiner: $malign->serializeOUT( "$wrkDir/$consName-malign-$round.ser"
)
./RepeatClassifier:# wublastx the simple-masked consensi vs the transposable ele
ment
./RepeatModeler:my $dbIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $genomeD
B`;
./RepeatModeler: my $sampleIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $s
ampleFastaFile`;
./RepeatModeler: # . " -mincount 3 -tandemdist 500 -output $fastaFile.lfreq
"
./RepeatModeler:"$workDir/sampleDB-$round.fa $workDir/consensi.fa -warnings -kap
-wordmask=dust wordmask=seg maskextra=10 -hspmax=20 V=0 B=250 Q=25 R=5 W=7 S=25
0 gapS2=250 S2=125 X=250 gapX=500 -matrix=comparison.matrix"
I am still waiting for a response...
I went to the web page of RepeatMasker that directs me to an array of other dependencies to be installed first. RepeatMasker depends on a repeat library RepBase which can be obtained from http://www.girinst.org. You go there and have to get yourself registered. Your registration need to be accepted by the group and then they send you an email carrying your login id and password. This may take upto 2 working days. Only after this you can download the RepBase database.
However, the trouble with RepBase is, it is a predefined library and will not detect any new repeat that may be there in your genome. So, the best option is to run a "De Novo" repeat finder. From the same web site you can get a link to RepeatModeler that acts as a wrapper around some de novo repeat finders such as RepeatScout and RECON.
Fair enough, then you go to install RepeatModeler...
This web page indicates you have to have a number of dependencies:
-- Of course you need to have perl 5.8 or above installed first.
-- you will need the Trfinder that I was mentioning earlier.
-- then you will need to download and install repeatmasker. This will not install unless you have RepBase in place.
--For RepeatMasker, you will also need phred/phrap/cross_match package. This can be done by writing to the authors and getting a copy by email. This may take a few business days.
--After you are done with repeatmasker you will need RECON to be installed
--Then RepeatScout need to be downloaded and installed.
--You will need WUBLAST also to be installed. The recommended site from the repeatmodeler page points to WUBLAST site at http://blast.wustl.edu/. When you go there you get redirected to http://www.advbiocomp.com . In advbiocomp site, they say that wublast is no longer supported and you can get an older version of WUblast for free. Then you get the binaries, inflate and keep them in place.
Now the final part: Installation of RepeatModeler. You do a ./configure. It asks you perl path, repeatMasker path etc. When it comes to WuBlast path, it exits saying that you xdformat program is older than 2005. What the hell, you back and check in wublast site, that says the program is as old as 1998. There are no newer version. There you get stuck.
Out of frustartion, I went and looked at the configure file in RepeatModeler. In line 237 there is a statement:
else {
my $result = `$wuLocation/xdformat -m 2>&1`;
# Two responses are expected:
# Good Version:
# /blah/wublast/xdformat: option requires an argument -- m
# Old Version:
# /blah/wublast/xdformat: invalid option -- m
unless ( $result =~ /option requires an argument -- m/ ) {
die "$wuLocation/xdformat is too old. Must have one dated 3/16/
5 "
. "or newer. Install a newer version of wublast and re-run
. "configure.\n";
}
I went back and asked the authors of xdformat, what this -m option means, they said to ignore it. Now I can change this, if only I find what is the substitute of this. Because my search in repeatModeler directory says me -m2 option is used:
gus@tyler-lab:~/RepeatModeler$ grep --regex=-m -r .
./configure: my $result = `$wuLocation/xdformat -m 2>&1`;
./Refiner: $malign->serializeOUT( "$wrkDir/$consName-malign-$round.ser"
)
./RepeatClassifier:# wublastx the simple-masked consensi vs the transposable ele
ment
./RepeatModeler:my $dbIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $genomeD
B`;
./RepeatModeler: my $sampleIndex = `$RepModelConfig::XDFORMAT_PRGM -m2 -n -r $s
ampleFastaFile`;
./RepeatModeler: # . " -mincount 3 -tandemdist 500 -output $fastaFile.lfreq
"
./RepeatModeler:"$workDir/sampleDB-$round.fa $workDir/consensi.fa -warnings -kap
-wordmask=dust wordmask=seg maskextra=10 -hspmax=20 V=0 B=250 Q=25 R=5 W=7 S=25
0 gapS2=250 S2=125 X=250 gapX=500 -matrix=comparison.matrix"
I am still waiting for a response...
Sunday, January 04, 2009
Author List
Recently there was a post by physioprof on the co-first author that appears second in the list. While I don't agree to with views that second listed first author to be completely ignored, but it does raise some question about authorship in papers.
While the younger lot generally go for quantity over quality and try to get themselves in any paper they find getting out of the group, many older un-productive ones that have been lying dormant for a long time get into papers through their experience. In many instances there are at least 30% of the people in a multi author paper get a free ride. There are also few others who generally get a ride through their influence or by having a good relationship with the PI. I know atleast a bunch of people who see their names in the paper by periodically checking their previous PI's website. They have no idea how and when the paper get published. Now this raises a serious question on genuine authorships in papers. In many journals there is a section to mention which author did what to the paper and I think that should be the norm
While the younger lot generally go for quantity over quality and try to get themselves in any paper they find getting out of the group, many older un-productive ones that have been lying dormant for a long time get into papers through their experience. In many instances there are at least 30% of the people in a multi author paper get a free ride. There are also few others who generally get a ride through their influence or by having a good relationship with the PI. I know atleast a bunch of people who see their names in the paper by periodically checking their previous PI's website. They have no idea how and when the paper get published. Now this raises a serious question on genuine authorships in papers. In many journals there is a section to mention which author did what to the paper and I think that should be the norm
Friday, January 02, 2009
Author List and Order
Recently there was a post by physioprof on the co-first author that appears second in the list. While I don't agree to his views that second listed first author to be completely ignored, but it does raise some question about authorship in papers.
While the younger lot generally go for quantity over quality and try to get themselves in any paper they find getting out of the group, many older un-productive ones that have been lying dormant for a long time
While the younger lot generally go for quantity over quality and try to get themselves in any paper they find getting out of the group, many older un-productive ones that have been lying dormant for a long time
Subscribe to:
Comments (Atom)
